on
Comparison of different for loops methods in parallel distance computation with R, C, and OpenMP
The title is a bit long, but I’ll try to keep the post short. In my previous post,
I presented how to convert a linear index to lower triangular subscripts. As
an example, it can be useful for distance computation. Rather than using two loops
for each row being compared in the input matrix, it is possible to use a single loop on an index,
and then compute the indices of the evaluated rows.
In a standard parallel distance implementation using two loops, all the computations
are done for the first row, then the second, …
I once heard that it could lead to a concurrency issue and limit the performance.
Using an index, we can proceed with a ‘diagonal-wise’ numbering, the combinations being computed in
parallel are then more likely to involved different rows, and this may consequently limit a putative concurrency
phenomenon. That’s what I aimed to check in this article.
The pmdr package
I wrote a toy R package, named pmdr for Parallel Manhattan Distances in R.
I focused on manhattan distances because it was easier to implement for this
exploratory approach. For simplicity, it only works with integers
and missing values are not allowed. Also, the results are returned as a vector.
This package contains three
C functions,
and the R wrapper.
It can be installed from Github as shown below.
# install.packages("remotes")
remotes::install_github("Atrebas/pmdr")Looping strategies
Using two loops
In the first C function, the loop is performed in a ‘classic’ way, with a
loop on i1 and i2, the rows being evaluated. From i1 and i2, k,
the index of the combination being evaluated is computed.
I is the number of rows of the input matrix ptx and ptres is the output
vector.
Note that the loop on j corresponds to the columns of the input matrix.
for (i1 = 1; i1 < I; i1++) { // loop on row 1
for (i2 = 0; i2 < i1; i2++) { // loop on row 2
k = K - (I - i2) * ((I - i2) - 1) / 2 + i1 - i2 - 1; // compute the index
ptres[k] = 0;
for (j = 0; j < J; j++) { // loop on the columns
ptres[k] += abs(ptx[i1 + I * j] - ptx[i2 + I * j]); // increment the Manhattan distance
}
}
}Using one loop, colwise
In the second C function, a loop is performed on k, the index of the
combination being evaluated. Within this loop, i1 and i2 are
determined in a colwise way.
for (k = 0; k < K; k++) { // loop on the index (column-wise)
ptres[k] = 0;
kp = K - k - 1;
p = floor((sqrt(1 + 8 * kp) - 1) / 2);
i1 = I - (kp - p * (p + 1) / 2) - 1; // compute row 1
i2 = I - 2 - p; // compute row 2
for (j = 0; j < J; j++) { // loop on the columns
ptres[k] += abs(ptx[i1 + I * j] - ptx[i2 + I * j]); // increment the Manhattan distance
}
}Using one loop, diagwise
In the third function, the parallelization is done on the linear index k, but
this time, the combination of rows used for the computation is determined
using diagwise traversal (and the order of the results in the output vector
will thus be different from the previous ones).
for (k = 0; k < K; k++) { // loop on the index (diagonal-wise)
ptres[k] = 0;
kp = K - k - 1;
p = floor((sqrt(1 + 8 * kp) - 1) / 2);
i1 = I - (kp - p * (p + 1) / 2) - 1; // compute row 1
i2 = i1 - (I - 1 - p); // compute row 2
for (j = 0; j < J; j++) { // loop on the columns
ptres[k] += abs(ptx[i1 + I * j] - ptx[i2 + I * j]); // increment the Manhattan distance
}
}Example
First, here is how the R function works. As mentioned earlier, I was interested in returning the results as a vector, not a distance matrix.
# remotes::install_github("atrebas/pmdr")
library(pmdr)
n_row <- 7
n_col <- 10
mat <- matrix(sample(1:30, n_row * n_col, replace = TRUE),
nrow = n_row,
ncol = n_col)
mat## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
## [1,] 6 17 26 23 15 20 13 11 1 13
## [2,] 25 28 1 12 18 1 10 9 10 12
## [3,] 13 19 16 14 4 16 7 24 8 30
## [4,] 24 22 16 12 20 16 3 1 24 10
## [5,] 8 11 11 21 23 4 21 21 7 25
## [6,] 25 10 5 24 2 16 11 16 16 12
## [7,] 10 3 2 30 22 7 23 8 18 24distance(mat, loop = "standard")## [1] 103 86 99 85 88 110 105 70 100 79 103 95 79 82 124 126 87 123 97
## [20] 59 94distance(mat, loop = "colwise")## [1] 103 86 99 85 88 110 105 70 100 79 103 95 79 82 124 126 87 123 97
## [20] 59 94distance(mat, loop = "diagwise")## [1] 103 105 95 126 97 94 86 70 79 87 59 99 100 82 123 85 79 124 88
## [20] 103 110Benchmark 1
Running a quick benchmark on my four-core laptop shows no benefit of the colwise and diagwise methods over the basic double loop.
library(pmdr)
library(microbenchmark)
n_row <- 500
n_col <- 1000
mat <- matrix(sample(1:30, n_row * n_col, replace = TRUE),
nrow = n_row,
ncol = n_col)
pmdr::num_procs()## [1] 4microbenchmark(standard = distance(mat, loop = "standard"),
colwise = distance(mat, loop = "colwise"),
diagwise = distance(mat, loop = "diagwise"),
times = 20L)## Unit: milliseconds
## expr min lq mean median uq max neval
## standard 214.3665 217.3569 219.9808 219.4761 223.6465 226.3603 20
## colwise 245.2472 245.9105 247.5465 246.4485 246.8566 266.0562 20
## diagwise 244.0165 250.4270 252.1025 251.4929 252.5880 275.2310 20Benchmark 2
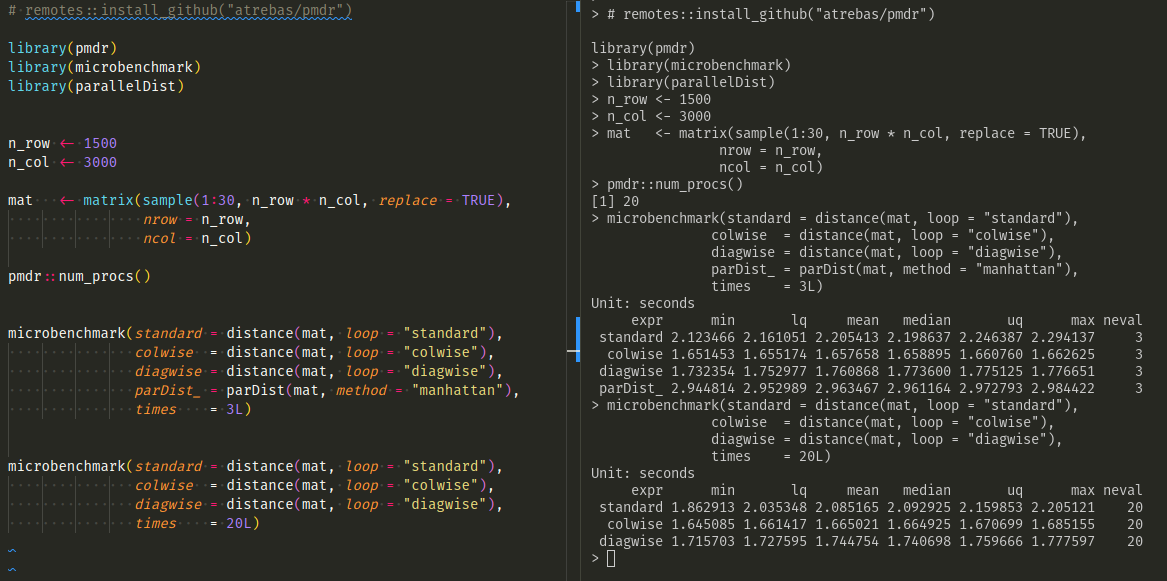
Here is a screenshot of a second benchmark with larger data
on a bigger machine (20 cores). In this case, the colwise method seems
to perform slightly better, but the benefit would probably vanish if a matrix
was returned (because in that case, computing the index in the double
loop would be useless).
Also, a quick test with the parallelDist package shows that the timing
obtained with pmdr is competitive.
Conclusion
No significant difference was observed between the different methods,
but writing the pmdr package was a good refresher on R/C binding and
my implementation of distance computation in OpenMP seems to work correctly.
The parallelDist package is much more robust and feature-rich than pmdr, so
the comparison is not relevant. Nevertheless, the timing obtained
in the benchmark above looks satisfying.
Note: I do not have a CS background. This post is based on MOOCs, reading, inspiration from other people’s work (Drew Schmidt’s Romp package in particular), and a lot of segfault debugging.